Biology Notes for Class 12
Chapter 6: Molecular Basis of Inheritance
Chapter Summary
Ø Nucleic acids are long polymers of nucleotides. While DNA stores genetic information, RNA mostly helps in transfer and expression of information. Though DNA and RNA both function as genetic material, but DNA being chemically and structurally more stable is a better genetic material. However, RNA is the first to evolve and DNA was derived from RNA. The hallmark of the double stranded helical structure of DNA is the hydrogen bonding between the bases from opposite strands. The rule is that Adenine pairs with Thymine through two H-bonds, and Guanine with Cytosine through three H-bonds. This makes one strand complementary to the other. The DNA replicates semiconservatively, the process is guided by the complementary H-bonding. A segment of DNA that codes for RNA may in a simplistic term can be referred as gene. During transcription also, one of the strands of DNA acts a template to direct the synthesis of complementary RNA. In bacteria, the transcribed mRNA is functional, hence can directly be translated. In eukaryotes, the gene is split. The coding sequences, exons, are interrupted by non-coding sequences, introns. Introns are removed and exons are joined to produce functional RNA by splicing. The messenger RNA contains the base sequences that are read in a combination of three (to make triplet genetic code) to code for an amino acid. The genetic code is read again on the principle of complementarity by tRNA that acts as an adapter molecule. There are specific tRNAs for every amino acid. The tRNA binds to specific amino acid at one end and pairs through H-bonding with codes on mRNA through its anticodons. The site of translation (protein synthesis) is ribosomes, which bind to mRNA and provide platform for joining of amino acids. One of the rRNA acts as a catalyst for peptide bond formation, which is an example of RNA enzyme (ribozyme). Translation is a process that has evolved around RNA, indicating that life began around RNA. Since, transcription and translation are energetically very expensive processes, these have to be tightly regulated. Regulation of transcription is the primary step for regulation of gene expression. In bacteria, more than one gene is arranged together and regulated in units called as operons. Lac operon is the prototype operon in bacteria, which codes for genes responsible for metabolism of lactose. The operon is regulated by the amount of lactose in the medium where the bacteria are grown. Therefore, this regulation can also be viewed as regulation of enzyme synthesis by its substrate.
Ø Human genome project was a mega project that aimed to sequence every base in human genome. This project has yielded much new information. Many new areas and avenues have opened up as a consequence of the project. DNA Fingerprinting is a technique to find out variations in individuals of a population at DNA level. It works on the principle of polymorphism in DNA sequences. It has immense applications in the field of forensic science, genetic biodiversity and evolutionary biology.
6.1 THE DNA:
Ø DNA is a long polymer of deoxyribonucleotides.
Ø The length of the DNA depends on, number of nucleotide pair present in it.
Ø Characteristics of the organism depend on the length of the DNA.
Ø Bacteriophage ø174 has 5386 nucleotides.
Ø Bacteriophage lambda has 48502 base pairs.
Ø Escherichia coli have 4.6 X 106 base pairs.
Ø Human genome (haploid) is 3.3 X 109 bp.
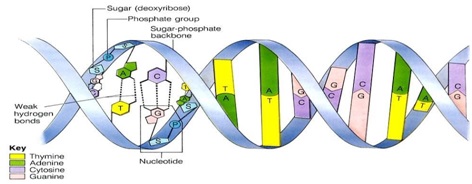
.1.1 Structure of polynucleotide chain:
Ø A nucleotide has three component:-
- A nitrogen base
- A pentose sugar ( ribose in RNA and deoxyribose in DNA)
- A phosphoric acid.
Ø There are two types of nitrogen bases:
- Purines ( Adenine and Guanine)
- Pyrimidines ( Cytosine, Uracil and Thymine)
Ø Adenine, Guanine and Cytosine is common in RNA and DNA.
Ø Uracil is present in RNA and Thymine is present in DNA in place of Uracil.
Ø Pentose sugar is ribose in RNA and Deoxyribose in DNA.
Ø A nitrogen base attached to the pentose sugar at C1 of pentose sugar by
Ø N-glycosidic linkage to form a nucleoside.
Ø According to the nature of pentose sugar, two types of nucleosides are formed ribonucleoside and deoxyribonucleotides.
Ø Ribonucleosides are:
- Adenosine
- Guanosine
- Cytidine
- Uridine
Ø Deoxyribonucleosides are:
- Deoxyadenosine
- Deoxyguanosine
- Deoxycytidine
- Deoxythymidine.
Ø Phosphoric acid attached to the 5’ OH of a nucleoside by Phosphodiester linkage a corresponding nucleotide is formed. (Ribonucleotide or deoxyribonucleotides depending on the sugar unit).
Ø Two nucleotides are joined by 3’-5’ Phosphodiester linkage to form dinucleotide.
Ø More than two nucleotides joined to form polynucleotide chain.
Ø Polynucleotide chain has a free phosphate moiety at 5’ end of sugar, is referred to as 5’ end
Ø In the other end of the polymer with 3’-OH group called 3’ end.
Ø The backbone of the polynucleotide chain is sugar and phosphate.
Ø Nitrogen bases linked to the sugar moiety project from the backbone.
Ø In RNA every nucleotide has an additional –OH group at 2’ of ribose.
Ø In RNA Uracil is found in place of thymine.
Ø 5-methyl uracil is the other name of thymine.
History of DNA:
Ø DNA is an acidic substance in the nucleus was first identified by Friedrich Meischer in 1869. He named it as ‘Nuclein”
Ø 1953 double helix structure of DNA was given by James Watson and Francis Crick, based on X- ray defraction data produced Maurice Wilkins and Rosalind Franklin.
Ø Hallmark of their proposition was base pairing between two strands of polynucleotide chains. This was based on observation of Erwin Chargaff.
Ø Chargaff’s observation was that for a double stranded DNA, the ratio between Adenine and Thymine, and Guanine and Cytosine are constant and equal one.
Salient features of Double helix structure of DNA:
Ø Made of two polynucleotide chains.
Ø Sugar and phosphate forms the backbone and bases projected to inside.
Ø Two chains have anti-parallel polarity.
Ø Two strands are held together by hydrogen bond present in between bases.
Ø Adenine of one strand pairs with Thymine of another strand by two hydrogen bonds and vice versa.
Ø Guanine of one strand pairs with Cytosine of another strand by three hydrogen bonds and vice versa.
Ø A purine comes opposite to a pyrimidine. This generates approximately uniform distance between the two strands of the helix.
Ø The two chains are coiled in a right – handed fashion.
Ø The pitch of the helix is 3.4 nm or 34 A°
Ø There are roughly 10 bp in turn.
Ø The distance between the bp in a helix is 0.34nm or 3.4 A°.
Ø The plane of one base pair stacks over the other in double helix.
Ø H-bond confers stability of the helical structure of the DNA.
Ø Central dogma of flow of genetic information: DNA→ RNA→ Protein.
6.1.2 Packaging of DNA Helix:
Ø Distance between two conjugative base pairs is 0.34nm, the length of the DNA in a typical mammalian cell will be 6.6 X109 bp X 0.34 X10-9 /bp, it comes about 2.2 meters.
Ø The length of DNA is more than the dimension of a typical nucleus (10-6m), how is such a long polymer packaged in a cell?
Packaging in prokaryotes:
Ø They do not have definite nucleus.
Ø The DNA is not scattered throughout the cell.
Ø DNA is held together with some proteins in a region is called ‘nucleoid’.
Ø The DNA in nucleoid is organized in large loops held be proteins.
Packaging in prokaryotes:
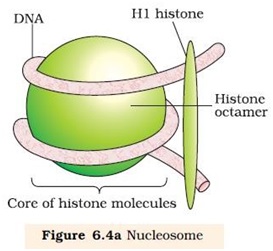
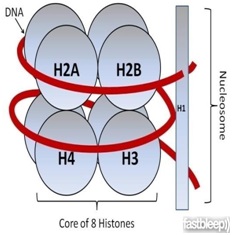
Ø In eukaryotes the packaging is more complex.
Ø There is a set of positively charged, basic protein called Histones.
Ø Histones are positively charged due to rich in basic amino acids like Lysines and arginines.
Ø Histones are organized to form a unit of eight molecules called histone octamere.
Ø Negatively charged DNA wrapped around positively charged histone octamere to form a structure called nucleosome.
Ø A typical nucleosome contains 200 bp of DNA helix.
Ø Nucleosome constitutes the repeating unit of a structure in nucleus called chromatin, thread like stained bodies seen in the nucleus.
Ø The nucleosomes are seen as ‘beads-on-string’ structure when viewed under electron microscope.
Ø The chromatin is packaged to form chromatin fibers that are further coiled and condensed at metaphase stage to form chromosome.
Ø Packaging at higher level required additional set of proteins called Non-histone Chromosomal (NHC) proteins.
Ø In a typical nucleus some loosely coiled regions of chromatin (light stained) is called euchromatin.
Ø The chromatin that more densely packed and stains dark are called Heterochromatin.
Ø Euchromatin is transcriptionally active chromatin and heterochromatin is inactive.
6.2 THE SEARCH OF GENETIC MATERIAL:
Transforming principle:
Ø Given by Frederick Griffith in 1928.
Ø His experiment based on Streptococcus pneumoniae (caused pneumonia).
Ø There is change in physical form of bacteria.
Ø There are two colonies of bacteria:
- Smooth shiny colonies called S strain.
- Rough colonies called R strain.
Ø S-strain bacteria have a mucous (polysaccharide) coat.
Ø R-strain does not have mucous coat.
Ø S-strain is virulent and caused pneumonia in mice and died when infected.
Ø R-strain is non-virulent and does not caused pneumonia in mice when infected.
Ø Heat killed S-Strain is non-virulent and does not causes pneumonia.
Ø The heat killed S-Strain mixed with live R-Strain injected into mice; the mice developed pneumonia and died.
Ø He recovered live S-Strain bacteria form the dead mice.
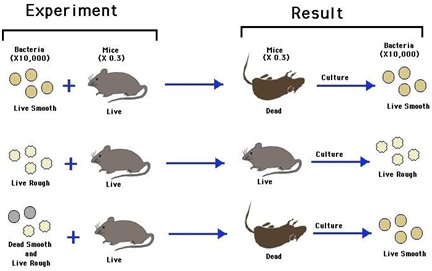
Conclusion of experiment:
Ø R – Strain bacteria had somehow been transformed by the heat killed S-Strain bacteria.
Ø Some ‘transforming principle’, transferred from heat killed S-Strain bacteria, had enabled the R-Strain to synthesize smooth polysaccharide coat and become virulent (S Strain).
Ø The transformation of R-Strain to S-Strain is due to transfer of Genetic material.
Ø However the biochemical nature of genetic material was not defined from his experiment.
Biochemical characterization of transforming principle:
Ø Biochemical nature of transforming principle was discovered by Oswald Avery, Colin Macleod and Maclyn McCarty. (1933-44)
Ø Prior to their work genetic material was thought to be protein.
Ø They worked to determine the biochemical nature of the ‘transforming principle’ of Griffith’s experiment.
Ø They purified biomolecules (proteins, DNA and RNA) from the heat killed S cells to see which one could transform live R cells to S cells.
Ø Heat killed S-Strain + protease + Live R-Strain → transformation.
Ø Heat killed S-Strain + RNase + Live R-Strain → transformation.
Ø Heat killed S-Strain + DNase + Live R-Strain → No transformation.
Conclusion of the experiments:
Ø Protein of heat killed S-Strain is not the genetic material
Ø RNA of heat killed S-Strain is not the genetic material.
Ø DNA of heat killed S-Strain is the genetic material, because DNA digested with DNase mixed with R-strain unable to transform R-Strain to S-Strain.
6.2.1 The Genetic Material is DNA:
Ø ‘DNA is the genetic material’ is proved by Alfred Hershey and Martha Chase (1952).
Ø They worked on the virus that infects bacteria called bacteriophage.
Ø During normal infection the bacteriophage first attaches the bacteria cell wall and then inserts its genetic material into the bacterial cell.
Ø The viral genetic material became integral part of the bacterial genome and subsequently manufactures more virus particle using host machinery.
Ø Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
Experiment 🙁 blenders experiment)
Ø They grew some viruses on a medium having radioactive phosphorus and some others on medium having radioactive sulfur.
Ø Viruses grown in radioactive Phosphorus have radioactive DNA but not radioactive protein because Phosphorus present in DNA not in protein.
Ø Viruses grown in radioactive sulfur have radioactive protein not radioactive DNA because sulfur present in protein but not in DNA.
Ø Infection: radioactive phages were allowed to attach to E.coli bacteria; the phages transfer the genetic material to the bacteria.
Ø Blending: the viral coats were separated from the bacteria surface by agitating them in a blender.
Ø Centrifugation: The virus particles were separated from the bacteria by spinning them in a centrifuge machine.
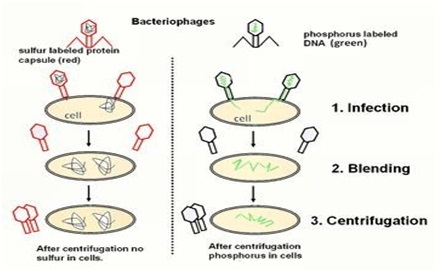
Observation:
Ø Bacteria infected with viruses that had radioactive DNA were radioactive and no radioactivity in the supernatant.
Ø Bacteria infected with viruses that had radioactive protein were not radioactive, but radioactivity found in the supernatant.
Conclusion of Experiment:
Ø DNA is the infecting agent that made the bacteria radioactive hence DNA is the genetic material not the protein.
6.2.2 Propoerties of Genetic Material (Dna Versus Rna):
Criteria for genetic material:
Ø It should be able to generate its replica (replication)
Ø It should be chemically and structurally stable.
Ø It should provide the scope for slow changes (mutation) that required for evolution.
Ø It should be able to express itself in the form of ‘Mendelian Character’.
Ø Protein dose not fulfill the criteria hence it is not the genetic material.
Ø RNA and DNA fulfill the criteria.
RNA is unstable:
Ø 2’-OH group present at every nucleotide (ribose sugar) in RNA is a reactive group and makes RNA liable and easily degradable.
Ø RNA is also now known as catalyst, hence reactive.
Ø RNA is unstable and mutates faster. Consequently the viruses having RNA genome and having shorter life span mutate and evolve faster.
DNA is more stable:
Ø Stability as one of the properties of genetic material was very evident in Griffith’s ‘transforming principle’ itself that heat, which killed the bacteria at least did not destroy some of the properties of genetic material.
Ø Two strands being complementary if separated by heating come together, when appropriate conditions are provided.
Ø Presence of Thymine in place of uracil confers additional stability to DNA
Ø DNA is chemically less reactive and structurally more stable when compared to RNA.
Ø Therefore among the two nucleic acids the DNA is a better genetic material.
Better genetic material (DNA or RNA)
Ø Presence of thymine at the place of uracil confers more stability to DNA.
Ø Both DNA and RNA are able to mutate.
Ø In fact RNA being unstable mutate at a faster rate.
Ø RNA can directly code for the synthesis of proteins, hence easily express.
Ø DNA however depends on RNA for protein synthesis.
Ø The protein synthesis machinery has evolved around RNA.
Ø Both RNA and DNA can functions as genetic material, but DNA being more stable is preferred for storage of genetic information.
Ø For the transmission of genetic information RNA is better.
6.3 RNA WORLD:
Ø RNA is the first genetic material.
Ø Essential life processes evolved around RNA.
Ø RNA used to act as a genetic material as well as catalyst.
Ø But RNA being catalyst was reactive and hence unstable.
Ø Hence DNA has evolved from RNA with chemical modifications that make it more stable.
Ø DNA being double stranded and having complementary strand further resists changes by evolving a process of repair.
6.4 REPLICATION: THE PROCESS:
Ø Watson and Crick proposed a scheme for replication of DNA.
Ø The Original statement that “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material (Watson and Crick, 1953)
Ø The scheme suggested that the two strands would separate and act as template for the synthesis of new complementary strands.
Ø New DNA molecule must have one parental strand and one new strand.
Ø This scheme of replication is called Semiconservative type of replication.
6.4.1 Experimental Proof of semiconservative nature of replication:
Ø It is now proved experimentally that replication is semiconservative type.
Ø It was first shown in Escherichia coli and subsequently in higher organism.
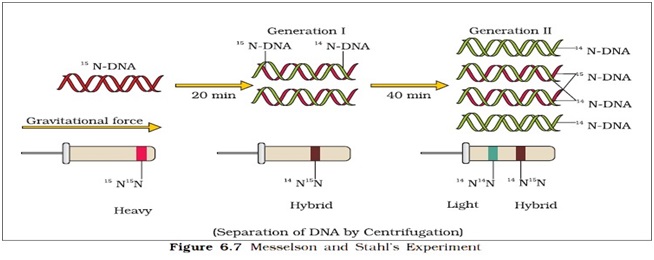
Ø Mathew Messelson and Franklin Stahl performed the following experiment in 1958.
Steps of the Experiments:
Ø They grew E.coli in 15NH4Cl medium for many generations. (15N is heavy nitrogen not radioactive element)
Ø The result was that 15N was incorporated into newly synthesized DNA and other nitrogen containing compound as well.
Ø This heavy DNA molecule could be distinguished from normal DNA by centrifugation in a cesium chloride (CsCl) density gradient.
Ø Then they transferred the E.coli into a medium with normal 14NH4Cl and let them grow.(E.coli divides in 20 minutes)
Ø They took samples at definite time intervals as the cells multiplied, and extracted the DNA that remained as double-stranded helices.
Ø Various samples were separated independently on CsCl gradients to measure the densities of DNA.
Ø The DNA that was extracted from the culture one generation after the transfer from 15N to 14N medium had a hybrid or intermediate density.
Ø DNA extracted from the culture after another generation (after 40 min.) was composed of equal amount of this hybrid DNA and of ‘light ‘DNA.
Experiment by Taylor and colleagues:
Ø Used radioactive thymidine to detect distribution of newly synthesized DNA in the chromosomes.
Ø They performed the experiment on Vicia faba (faba beans) in 1958.
Ø They proved the semiconservative nature of DNA replication in eukaryotes.
6.4.2 Replication Machinery and Enzymes:
Ø In all living cells such as E.coli replication requires a set of enzymes.
Ø E.coli completes the replication of its DNA in within 38 min.
Ø The average rate of polymerization has to be approx. 2000 bp per sec.
Ø The polymerization process must be accurate; any mistake during replication would result into mutation.
Ø Deoxyribonucleoside triphosphates (dATP, dGTP, dCTP, dTTP) serve dual purposes:
Ø Provide energy for polymerization.
Ø Acts as substrates for polymerization.
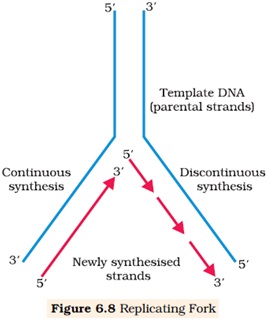
Ø The replication process occurs within a small opening of the DNA helix called replication fork.
Ø The region where, replication fork formed is called origin of replication.
Ø The replication fork is formed by an enzyme called helicase.
Ø Two separated strand is called template strands.
Ø Main enzyme is DNA-dependent DNA polymerase, since it uses a DNA template to catalyze the polymerization of deoxyribonucleotides.
Ø DNA polymerase catalyses polymerization only in one direction i.e. 5’→3’.
Ø On one strand (template with 3’→5’ polarity) the replication is continuous hence called leading strand.
Ø In another strand (template with 5’→3’ polarity) the polymerization takes place in the form of short fragment called Okazaki fragment.
Ø The short fragments are joined by DNA ligase, hence called lagging strand.
Ø In eukaryotes replication takes place in S-phase of cell cycle.
Ø A failure of cytokinesis after replication results into polyploidy.
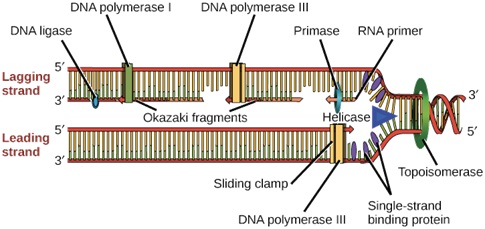
Enzyme used I DNA Replication process-
Ø Helicase opens up the DNA at the replication fork.
Ø Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
Ø Topoisomerase works at the region ahead of the replication fork to prevent supercoiling.
Ø Primase synthesizes RNA primers complementary to the DNA strand.
Ø DNA polymerase III extends the primers, adding on to the 3′ end, to make the bulk of the new DNA.
Ø RNA primers are removed and replaced with DNA by DNA polymerase I.
Ø The gaps between DNA fragments are sealed by DNA ligase.
6.5 TRANSCRIPTION:
Ø ‘The process of copying genetic information from one strand of the DNA into RNA is termed as transcription’.
Ø Transcription vs. Replication:
- Principle of complementarity governs the process of transcription except Adenosine of DNA forms base pair with theUracil instead of thymine. During replication Adenine pairs with thymine instead of uracil.
- During replication once started the whole DNA is duplicated, where as transcription takes place only a segment of DNA.
- In replication both strand acts as template, where as in transcription only one strand is acts as template to synthesize RNA.
- In replication DNA copied from a DNA, where as in transcription RNA copied from the DNA.
Why both strands of DNA not copied during transcription:
Ø If both strand of DNA acts as template, they would translated into two RNA of different sequences and in turn if they code for proteins, the sequence of amino acids in the protein would be different. Hence one segment of DNA would be coding for two different proteins.
Ø The two RNA molecules if produced from simultaneously would be complementary to each other, hence will form double stranded RNA. This would prevent RNA translation into protein.
6.5.1 Transcription unit:
Ø A transcription unit in DNA consists of three regions:
Ø A promoter
Ø The structural gene
Ø A terminator.
Ø DNA dependent RNA polymerase catalyses the polymerization in only one direction that is 5’→3’.
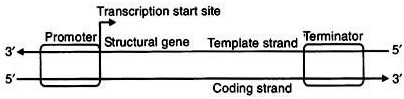
Structural gene:
Ø The DNA strand having polarity 3’→5’ is called template strand for transcription.
Ø The other strand of DNA having polarity 5’→3’ is called coding strand.
Ø The sequences of nitrogen base in the RNA transcribed from the template strand are same as the coding strand of DNA except having Thymine in place of Uracil.
Ø All the reference point defining a transcription unit is made with the coding strand only, not the template strand.
Promoter:
Ø Promoter and Terminator present on either side of structural gene.
Ø The promoter located towards 5’ end (upstream) of the structural gene.
Ø It is a short sequence of DNA that provides binding site for RNA polymerase. (mostly TATA , Commonly called TATA box)
Ø Presence of the promoter defines the template and coding strands.
Ø If the position of promoter is changed with terminator the definition of coding and template strand will be reversed.
Terminator:
Ø The terminator located towards 3’ end (down stream) of coding strand.
Ø It terminates the process of transcription.
Ø It is also a short segment of DNA which recognizes the termination factor. (ρ-factor)
6.5.2 Transcription unit and the gene:
Ø Gene is defined as the functional unit of inheritance.
Ø Genes are located on the DNA.
Ø The DNA sequence coding for tRNA and rRNA molecule also define a gene.
Ø Cistron: a segment of DNA (structural gene) coding for a polypeptide.
Ø Monocistronic: most of eukaryotic structural gene codes for single polypeptide.
Ø Polycistronic: Most prokaryotic structural gene code for more than one polypeptides.
Ø In eukaryotes the monocistronic structural gens have interrupted coding sequences, the genes are said to be split gene:
- The coding sequences or expressed sequences are called Exons.
- Exons are interrupted by Introns.
Ø Exons are said to be those sequences that appear in mature or processed mRNA.
Ø Introns never appear in mature of processed mRNA. They are spliced out.
6.5.3 Types of RNA:
Ø In prokaryotes there are three major types of RNAs: mRNA (messenger), tRNA (transfer), and rRNA (ribosomal).
Ø All three RNAs are required to synthesize protein in a cell.
Ø The mRNA provides the template and having genetic information in the form of genetic code.
Ø The tRNA brings the amino acids and read the genetic code of mRNA.
Ø The rRNA is the structural part of the ribosome and also as catalytic role during process of translation.
Process of transcription: prokaryotes:
Ø There is a single DNA dependent RNA polymerase that catalyses transcription or synthesis of all three types of RNAs in prokaryotes.
Ø The process of transcription completed in three steps:

Initiation:
Ø RNA polymerase binds to the specific site of DNA called promoter.
Ø Promoter of the DNA is recognized by initiation factor or sigma (σ).
Ø RNA polymerase along with initiation factor binds to the promoter.
Elongation:
Ø RNA polymerase unzipped the DNA double helix and forms an open loop.
Ø It uses ribonucleoside triphosphates as substrate and polymerizes in a DNA template following the rule of complementarity.
Ø Only a short stretch of polymerized RNA remains binds with the enzyme.
Ø The process of polymerization continued till the enzyme reaches the terminator gene.
Termination:
Ø RNA polymerase recognizes the terminator gene by a termination-factor called rho (ρ) factor.
Ø The RNA polymerase separated from the DNA and also the transcribed RNA.
Additional complexities in eukaryotes:
Ø There are three different types of RNA polymerases in the nucleus:
- RNA polymerase I transcribes rRNA (28S, 18S, and 5.8S)
- RNA polymerase II transcribes heterogeneous nuclear RNA (hnRNA).
- RNA polymerase III transcribes tRNA, 5srRNA and snRNA.
Ø Post transcriptional processing: (occurs inside the nucleus)
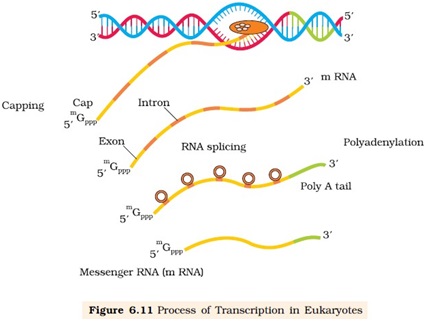
Splicing:
Ø The primary transcript (hn RNA) contain both exons and introns and required to be processed before translationally active (mRNA).
Ø The introns are removed and exons are joined in a defined order.
Ø This process is catalyzed by SnRNP, introns removed as spliceosome.
Capping: an unusual nucleotide called methyl guanosine triphosphate is added to the 5’ end of hnRNA.
Tailing: Adenylate residues (200-300) are added at 3’ end of hnRNA in a template independent manner.
The processed hnRNA is now called mRNA and transported out of the nucleus for translation.
6.6 GENETIC CODE:
Contribution to discovery:
Ø The process of replication and transcription based on complementarity.
Ø The process of translation is the transfer of genetic information form a polymer of nucleotides to a polymer of amino acids. There is no complementarity exist between nucleotides and amino acids.
Ø If there is change in the nucleic acid (genetic material) there is change in amino acids in proteins.
Ø There must be a genetic code that could direct the sequence of amino acids in proteins during translation.
Ø George Gamow proposed the code should be combination of bases, he suggested that in order to code for all the 20 amino acids, the code should be made up of three nucleotides.
Ø Har Govind Khorana enables instrumental synthesizing RNA molecules with desired combinations of bases (homopolymer and copolymers).
Ø Marshall Nirenberg’s cell – free system for protein synthesis finally helped the discovery of genetic code.
Ø Severo Ochoa enzyme (polynucleotide phosphorylase) was also helpful in polymerizing RNA with desired sequences in a template independent manner (enzymatic synthesis of RNA).
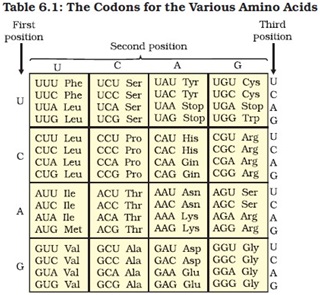
Salient features of genetic code:
Ø The codon is triplet. Three nitrogen base sequences constitute one codon.
Ø There are 64 codon, 61 codes for amino acids and 3 codons are stop codon.
Ø One codon codes for only one amino acid, hence it is unambiguous.
Ø Degeneracy: some amino acids are coded by more than one codon.
Ø Comma less: the codon is read in mRNA in a continuous fashion. There is no punctuation.
Ø Universal: From bacteria to human UUU codes for phenyl alanine.
Ø Initiation codon: AUG is the first codon of all mRNA. And also it codes for methionine (met), hence has dual function.
Ø Non-overlapping: The genetic code reads linearly
Ø Direction: the code only read in 5’ → 3’ direction.
Ø Anticodon: Each codon has a complementary anticodon on tRNA.
Ø Non-sense codon: UAA, GUA, and UAG do not code for amino acid and has no anticodon on the tRNA.
6.6.1 Mutation and Genetic code:
Ø Relationship between DNA and genes are best understood by mutation.
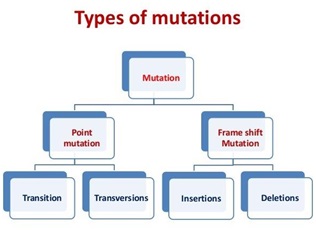
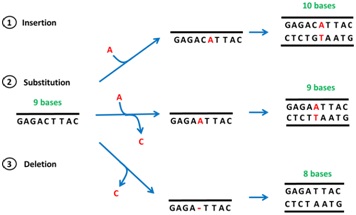
Point mutation:
Ø It occurs due to replacement nitrogen base within the gene.
Ø It only affects the change of particular amino acid.
Ø Best understood the cause of sickle cell anemia.
Frame shift mutation:
Ø It occurs due to insertion or deletion of one or more nitrogen bases in the gene.
Ø There is change in whole sequence of amino acid from the point of insertion or deletion.
Best understood in β-thalasemia.
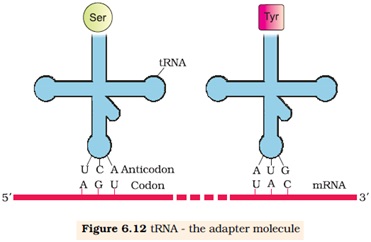
6.6.2 tRNA-the Adaptor molecule:
Ø The tRNA is called sRNA (soluble RNA)
Ø It acts as an adapter molecule.
Ø tRNA has an anticodon loop that base complementary to the codon.
Ø It has an amino acid accepter end to which it binds with amino acid.
Ø Each tRNA bind with specific amino acid i.e 61 types of tRNA found.
Ø One specific tRNA with anticodon UAC called initiator tRNA.
Ø There is no tRNA for stop codons. (UAA, UGA, UAG)
Ø The secondary structure is like clover-leaf.
Ø The actual structure of tRNA is compact, looks like inverted ‘L’.
6.7 TRANSLATION:
Ø It refers to polymerization of amino acids to form a polypeptide.
Ø The number and sequence of amino acids are defined by the sequence of bases in the mRNA.
Ø The amino acids are joined by peptide bond.
Ø Amino acids are activated in the presence of ATP and linked to their specific tRNA is called charging of tRNAor aminoacylation of tRNA.
Ø Ribosome is the cellular factory for protein synthesis.
Ø Ribosome consists of structural rRNA and 80 different proteins.
Ø In inactive state ribosome(70S) present in two subunits:-
- A large sub unit 50S.
- A small sub unit 30S

Initiation:
Ø The process of translation or protein synthesis begins with attachment of mRNA with small subunit of ribosome.
Ø The ribosome binds to the mRNA at the start codon (AUG).
Ø AUG is recognized by the initiator tRNA.
Elongation:
Ø Larger subunit attached with the initiation complex.
Ø Larger subunit has two site ‘A’ site and ‘P’ site.
Ø Initiator tRNA accommodated in ‘P’ site of large subunit, the subsequent amino-acyl-tRNA enters into the ‘A’ site.
Ø The sub subsequent tRNA selected according to the codon of the mRNA.
Ø Codon of mRNA and anticodon of tRNA are complementary to each other.
Ø Formation of peptide bond between two amino acids of ‘P’ and ‘A’ site, catalyzed by ribozyme, (23S rRNA in bacteria)
Ø The moves from codon to codon along the mRNA called translocation.
Termination:
Ø Elongation continues until a stop codon arrives at ‘P’ site.
Ø There is no tRNA for stop codon.
Ø A release factor binds to the stop codon.
Ø Further shifting of ribosome leads to separation of polypeptide.
Ø An mRNA also has some additional sequences that are not translated called untranslated regions (UTR).
6.8 REGULATION OF GENE EXPRESSION:
Ø Regulation of gene expression in eukaryotes takes place in different level:
Ø Transcriptional level (formation of primary transcript)
Ø Processing level (regulation of splicing)
Ø Transport of mRNA from nucleus to the cytoplasm.
Ø Translational level.
Ø In prokaryotes control of rate of transcriptional initiation is the predominant site for control of gene expression.
Ø The activity of RNA polymerase at the promoter is regulated by accessory proteins, which affects its ability to recognize the start site.
Ø The regulatory proteins can acts both positively (activators) or negatively (repressor)
Ø The regulatory proteins interact with specific region of DNA called operator, which regulate the accessibility of RNA polymerase to promoter.
Lac operon:
Ø Francois Jacob and Jacque Monod first to describe a transcriptionally regulated system of gene expression.
Ø A polycistronic structural gene is regulated by common promoter and regulatory genes. Such regulation system is common in bacteria and is called operon.
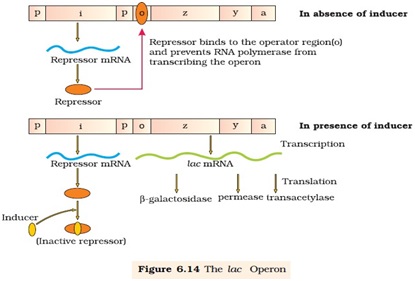
Ø Lac operon consists of:-
- One regulator gene ( i-gene)
- Three structural genes (z,y,a)
- Operator. (binding site of repressor protein)
- Promoter.(binding site of the RNA polymerase)
Ø The i-gene codes for repressor of the lac operon.
Ø The structural gene consist of three gene (z, y and a)
- ‘z’-gene codes for beta-galactosidase, which hydrolyze lactose into Galactose and glucose.
- ‘y’ –gene codes for permease, which increases the permeability of bacterial cell to lactose.
- ‘a’-gene codes for transacetylase.
Ø All three genes are required for the metabolism of lactose in bacteria.
Ø Inducer: lactose is the substrate for β- galactosidase and it regulates the switching on and off of the lac operon. Hence it is called inducer.
Ø In the absence of glucose, if lactose is added in the growth medium of the bacteria, the lactose is transported into the cell by permease.
Ø Very low level of expression of lac operon has to be present in the cell all the time; otherwise lactose cannot enter the cell.
Mechanism of regulation of lac operon:
Ø The repressor protein is synthesized from i-gene (all time constitutively)
Ø In the absence of the inducer i.e. lactose the active repressor binds to the operator and prevents RNA polymerase from transcribing the structural gene
Ø In the presence of the inducer such as lactose or allolactose, the repressor is inactivated by interaction with inducer.
Ø This allows RNA polymerase access to the promoter and transcription proceeds.
Ø The regulation of lac operon by repressor is referred to as negative regulation.
6.9 HUMAN GENOMIC PROJECT:
Ø Genetic make-up of an organism or an individual lies in the DNA sequences.
Ø Two individual differs in their DNA sequences at least in some places.
Ø Finding out the complete DNA sequence of human genome.
Ø Sequencing human genome was launched in 190.
Goals of HGP:
Ø Identify all the approximately 20.000 – 25000 genes in human DNA.
Ø Determine the sequence of all 3 billion chemical base pairs.
Ø Store this information in data bases.
Ø Improve tools for data analysis.
Ø Transfer related technologies to other sectors, such as industries.
Ø Address the ethical, legal, and social issues (ELSI) that may arise from the project.
Methodology:
Ø To identify all the genes that expressed as RNA referred as Expressed Sequence Tags (ETSs).
Ø Simply sequencing the whole set of genome that contained all the coding and non-coding sequence, and later assigning different regions in the sequence with functions called Sequence Annotation.
Ø The commonly used hosts for sequencing were bacteria and yeast and vectors were called as BAC (bacterial artificial chromosome) and YAC (yeast artificial chromosome).
6.9.1 Salient features of Human Genome:
Ø The human genome contains 3164.7 million nucleotide bases.
Ø The average gene consists of 3000 bases.
Ø The largest known human gene being dystrophin at 2.4 million bases.
Ø The total number of gene is estimated at 30.000.
Ø 99.9 percent nucleotide base sequences are same in all peoples.
Ø The function of 50% genes discovered is unknown.
Ø Less than 2 percent of the genome codes for proteins.
Ø Repeated sequences make up very large portion of human genome.
Ø Chromosome I has most genes (2968) and the Y has the fewest (231).
Ø It is identified about 1.4 million locations where single-base DNA differences (SNPs – single nucleotide polymorphism) occurs in humans.
6.10 DNA FINGER PRINTING:
Ø DNA finger printing is a very quick way to compare the DNA sequences of any two individual.
Ø DNA fingerprinting involves identifying differences in some specific regions in DNA called repetitive DNA,because in these sequences, a small stretch of DNA is repeated many times.
Ø During centrifugation the bulk DNA forms major peak and the other small peaks are called satellite DNA.
Ø Depending on base composition (A:T rich or G:C rich), length of segment, and number of repetitive units, the satellite DNA classified into many types, such as mini –satellite and micro – satellite.
Ø These sequences dose not code for any proteins.
Ø These sequences show high degree of polymorphism and form basis of DNA fingerprinting.
Ø Polymorphism in DNA sequence is the basis of genetic mapping of human genome as well as of DNA fingerprinting.
Ø Polymorphism (variation at genetic level) arises due to mutations.
Ø If an inheritable mutation is observed in a population at high frequency it is referred as DNA polymorphism.
The process:
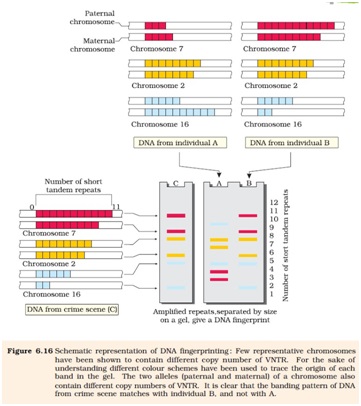
Ø DNA fingerprinting was initially developed by Alec Jeffreys.
Ø He used satellite DNA as the basis of DNA fingerprinting that shows very high degree of polymorphism. It was called as Variable Number Tandem Repeats.(VNTR)
Ø Different steps of DNA fingerprinting are:-
- Isolation of DNA.
- Digestion of DNA by restriction endonucleases.
- Separation of DNA fragments by gel electrophoresis.
- Transferring (blotting) of separated DNA fragments to synthetic membranes, such as nitrocellulose or nylon.
- Double stranded DNA made single stranded.
- Hybridization using labeled VNTR probe.
- Detection of hybridized DNA fragments by autoradiography.
Ø The VNTR belongs to a class of satellite DNA referred to as mini-satellite.
Ø The size of VNTR varies from 0.1 to 20 kb.
Ø After hybridization with VNTR probe the autoradiogram gives many bands of different sizes. These bands give a characteristic pattern for an individual DNA. It differs from individual to individual.
Ø The DNA from a single cell is enough to perform DNA fingerprinting.
Applications:
Ø Test of paternity.
Ø Identify the criminals.
Ø Population diversity determination.
Determination of genetic diversity
Disclaimer: All contents are originally prepared by Shri K C Meena Ji, Principal, KVS.
Amazon Affiliate Disclaimer: cbsecontent.com is a part of Amazon Services LLC Associates Program, an affiliate advertising program designed to provide a means for sites to earn advertising fees by advertising and linking to Amazon.in. As an amazon associates we earn from qualifying purchases.










